Batch Processing Using Jupyter and Python¶
Some data management or metadata tasks can best be tackled by writing a script to automate processes that are repetitive or repeated regularly. Since the Metadata Wizard is built on, and ships with a fairly complete Python stack, we allow advanced users to leverage this functionality using a Jupyter Notebook.
Prerequisites¶
Use of scripting, as described below, will require a basic knowledge of the Python programing language as well as how to use Jupyter Notebooks. There are many excellent resources for learning these powerful tools such as:
Launching Jupyter From the Metadata Wizard¶
A live instance of a Jupyter Notebook kernel can be launched directly from the Metadata Wizard application by clicking Launch Jupyter in the Advanced menu.
Menu item to Launch Jupyter¶
The user will be then need to specify what directory would like to open Jupyter in. This will be a project workspace or other folder containing the notebook files (.ipynb) they would like to run or edit. The default directory contains example notebooks that are described below. Choose Browse to to navigate to a specific project folder or specify one of the last 10 directories used from the dropdown box. If you have Anaconda installed and have set up environments with specific packages installed, you can choose one from the combobox below to use for this Jupyter session.
Click Launch to start Jupyter, which will appear momentarily in your default web browser.
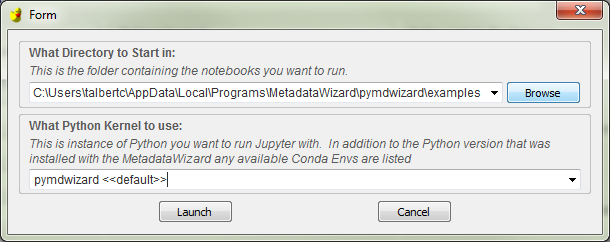
Prompt for choosing where to start the Notebook server and optionally which Anaconda env to use.¶
Example Notebooks¶
The Metadata Wizard ships with a set of example Notebooks that are intended to provide a demonstration of how one might use these capabilities. Each is self-documenting, in that it contains internal explanations of the code contained in it’s cells.
pymdwizard scripting (Start Here).ipynb – Provides an introduction to opening, searching, editing, saving, and validating FGDC metadata. It uses the pymdwizard’s core functionality and is probably where most users will want to start.
Report on all metadata in a directory.ipynb – Provides an example of how one might generate a report on all the metadata contained in a directory including the FGDC schema errors in each file.
FGDC ITIS Taxonomy generation.ipynb – Provides an example of how to use the core functionality to search the Integrated Taxonomic Information System (ITIS) by scientific and common name and generate FGDC taxonomy sections using code.
BatchUpdateAuthorsDatesEtc.ipynb – A use case for how the above techniques were used to update a large batch of existing metadata records.
MD_Propogation_and_SB_DataUpload.ipynb – A use case for how the above techniques were extended to generate metadata for multiple datasets and move the associated data and metadata up to an online repository (a data release on the USGS ScienceBase system).